Memory Usage of a Large NumPy Array
February 4, 2023
In the examples below, a 500x2000x2000 three-dimensional NumPy array named a is initialized with zeros. At each iteration, a random two-dimensional array r is inserted into array a. This approach could represent a larger piece of code where the r array would be created from various calculations during each iteration of the for-loop. Consequently, each slice in the z dimension of the a array is calculated at each iteration.
The examples were run on a MacBook Pro with a 2.6 GHz 6-Core Intel Core i7 CPU and 32 GB of memory (RAM). Memory usage is profiled with the memory-profiler tool using the terminal commands shown below.
# Generate a memory profile for ex1_basic.py
mprof run --output ex1_mprof.dat ex1_basic.py
mprof plot --output ex1_mprof.pdf ex1_mprof.dat
Example 1
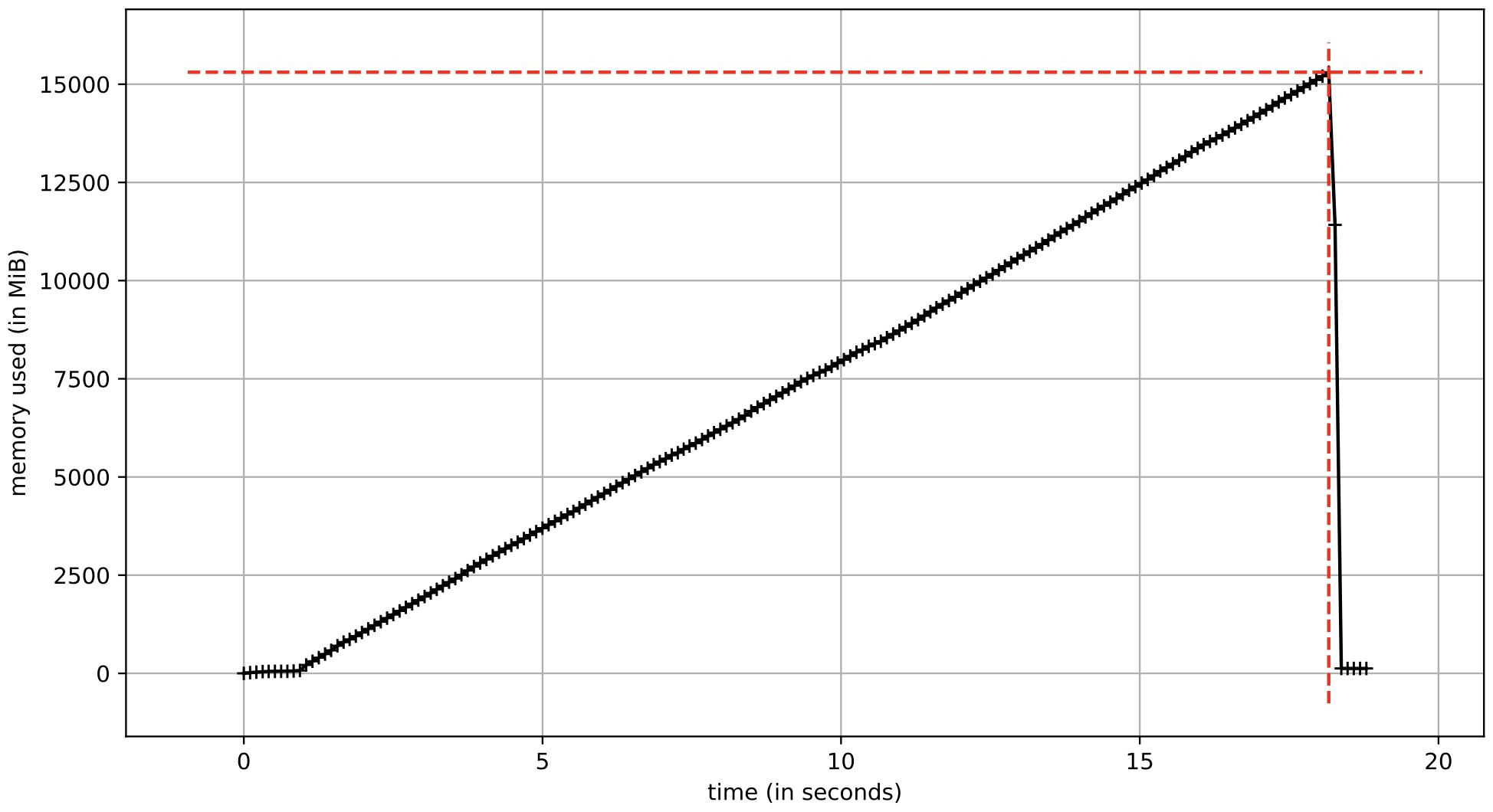
Basic example of building a large NumPy array with random values. Notice that np.random.rand() creates an array of np.float64 values.
# ex1_basic.py
import numpy as np
import time
def main():
tic = time.perf_counter()
z = 500 # depth
x = 2000 # rows
y = 2000 # columns
a = np.zeros((z, x, y))
for i in range(z):
r = np.random.rand(x, y)
a[i] = r
toc = time.perf_counter()
print('elapsed time =', round(toc - tic, 2), 'sec')
s = np.float64().nbytes * (z * x * y) / 1e9 # where 1 GB = 1000 MB
print('calculated storage =', s, 'GB')
if __name__ == '__main__':
main()
$ python ex1_basic.py
elapsed time = 16.56 sec
calculated storage = 16.0 GB
Example 2
An example of building a large NumPy array with random values where the data type is
defined as np.float32.
# ex2_float32.py
import numpy as np
import time
def main():
rng = np.random.default_rng()
tic = time.perf_counter()
z = 500 # depth
x = 2000 # rows
y = 2000 # columns
a = np.zeros((z, x, y), dtype=np.float32)
for i in range(z):
r = rng.standard_normal((x, y), dtype=np.float32)
a[i] = r
toc = time.perf_counter()
print('elapsed time =', round(toc - tic, 2), 'sec')
s = np.float32().nbytes * (z * x * y) / 1e9 # where 1 GB = 1000 MB
print('calculated storage =', s, 'GB')
if __name__ == '__main__':
main()
$ python ex2_float32.py
elapsed time = 19.41 sec
calculated storage = 8.0 GB

Example 3
Using the h5py package, this example creates an hdf5 file that contains a dataset that represents the a array. The dset variable is similar to the a variable in the previous examples. This approach allows the array to reside on disk, not in memory.
# ex3_hdf5.py
import numpy as np
import h5py
import time
def main():
rng = np.random.default_rng()
tic = time.perf_counter()
z = 500 # depth
x = 2000 # rows
y = 2000 # columns
f = h5py.File('file.hdf5', 'w')
dset = f.create_dataset('data', shape=(z, x, y), dtype=np.float32)
for i in range(z):
r = rng.standard_normal((x, y), dtype=np.float32)
dset[i, :, :] = r
toc = time.perf_counter()
print('elapsed time =', round(toc - tic, 2), 'sec')
s = np.float32().nbytes * (z * x * y) / 1e9 # where 1 GB = 1000 MB
print('calculated storage =', s, 'GB')
if __name__ == '__main__':
main()
$ python ex3_hdf5.py
elapsed time = 21.34 sec
calculated storage = 8.0 GB

Summary
A comparison of the three examples is shown below in the table. Changing the array data type from float64 to float32 cut the memory usage in half. Writing the array to disk drastically reduced memory use. The generated hdf5 file is about 8 GB on disk which is the size of the array containing float32 values. The elapsed time for the hdf5 approach is similar to the other examples; therefore, writing to the hdf5 file seems to have a negligible performance impact. Based on these examples, writing the array to disk using an hdf5 file allows substantial memory reduction as long as disk space is available for storing the array.
| Example | Elapsed time | Calculated storage | Peak memory | File storage |
|---|---|---|---|---|
| 1 basic | 16.56 s | 16.0 GB | 16.05 GB | n/a |
| 2 float32 | 19.41 s | 8.0 GB | 8.06 GB | n/a |
| 3 hdf5 | 21.34 s | 8.0 GB | 104 MB | 8.02 GB |
Other approaches for writing the array to disk such as numpy.memmap and zarr were tested too. But the hdf5 approach demonstrated in example 3 gave better performance and memory usage for this type of problem.
A comment about the memory increase over time for examples 1 and 2. When an array is initialized with np.zeros, the memory is lazily allocated as values are added to the array. The total amount of memory allocated for the array is based on the data type of the values stored in the array.
Pythonic Programming © 2025
Built with Genja by Gavin Wiggins